Non allelic homologous recombination (NAHR) is a key genetic process for creating variety.1 It happens when genetic material swaps between non-allelic, similar chromosomes. In this guide, we’ll look into the meaning, importance, how it works, and what it means for plants and people. NAHR can cause changes in chromosomes like deletions, duplications, and inversions. These changes are critical for keeping genomes stable and affecting human health.
Key Takeaways
- Non allelic homologous recombination (NAHR) is a genetic process that generates genetic diversity during meiosis.
- NAHR can lead to chromosomal rearrangements, including deletions, duplications, and inversions, which can impact genome stability and human health.
- NAHR has been extensively studied in plants, particularly in the context of soybean seed coat color.
- The repetitive nature of certain genomic regions poses challenges for genome assembly, which can be addressed using techniques like bacterial artificial chromosome (BAC) sequencing.
- NAHR-mediated chromosomal rearrangements have been linked to various genomic disorders in humans, including male infertility.
What is Non Allelic Homologous Recombination?
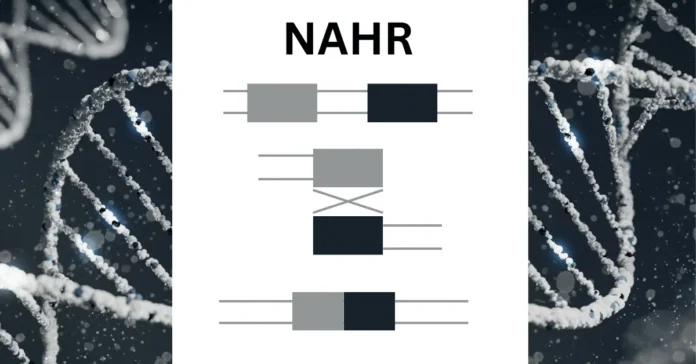
Non allelic homologous recombination (NAHR) is a complex type of genetic recombination. It occurs between DNA sequences that look alike but aren’t the same.2 They’re found in different places on chromosomes, or on different chromosomes entirely. During a specific part of cell division, called meiosis, NAHR swaps genetic material between these similar regions. This can cause chromosomes to change, by deleting, duplicating, or flipping sections.
Definition and Explanation
NAHR involves the cross-like swapping of genetic material kind of like a jigsaw mix-up. It happens between similar DNA sections, which can be quite long. These sections match up about 95-97% in sequence.2 Because they’re so alike, they can mistakenly recombine, leading to changes in the chromosomes’ structure.
Significance in Genetic Diversity
NAHR is a key player in shaking up genetic variation. It mixes up genes from non-identical places, adding unique genetic cocktails to populations.2 While this mostly boosts genetic diversity, it can sometimes flub up important genes, causing rare genetic issues.
| Metric | Value |
|---|---|
| Number of accesses to the research article | 98813 |
| Number of citations | 343 |
| Number of NAHR-mediated deletions or duplications identified in 44 individuals | 109 out of 324 potential NAHR loci3 |
| Percentage of genome rearrangements thought to be the result of NAHR in humans | approximately 10% to 22%3 |
| Proportion of NAHR breakpoints that are supported by ≤2 discordant paired-end reads | 93%3 |
| Number of individuals analyzed from the 1000 Genomes Project dataset | 443 |
| Number of called NAHR events detected in the analysis | 1,0433 |
| Median number of individuals per locus with a call for NAHR events | five3 |
| Availability of the algorithm detect-NAHR | freely accessible3 |
Non Allelic Homologous Recombination in Plants
Scientists have looked into non allelic homologous recombination (NAHR) a lot, especially in plants. This happens most in the soybean seed coat color. This color comes from the I locus which has many chalcone synthase (CHS) gene copies.4
Role in Soybean Seed Coat Color
In soybeans, the number of CHS gene copies changes with the seed coat’s color. Cultivars with different I alleles show this. Among them, ii types have the most copies, then I types, then ik.4 Spontaneous changes in i alleles reduce the copy number. This is shown by digital PCR comparing them to the original types.4
Structure of Chalcone Synthase (CHS) Gene Repeats
The CHS genes in the I locus have a special structure. They have two sets of the same genes, with a smaller DNA piece between them.4 Changes like NAHR can remove parts of the genes. This can lead to new seed coat colors.4 Special DNA regions can cause these changes, like cuts in the DNA, turning genes off, or adding new DNA.4
Genome Assembly Challenges
Soybean genome assembly work is hard due to repeating sections. The I locus area, with many CHS genes, is especially tricky. When matching resequenced data to current soybean assemblies, like Gmax109 Wm82.a1 and Gmax275 Wm82.a2, big gaps and swaps were found. This means the original assemblies didn’t fully show the I locus.4
Gaps and Inversions in Soybean Genome Assemblies
Genome assembly challenges get harder because of the repeating CHS gene blocks in the I locus. This area has a unique pattern: two sets of CHS genes are the same but arranged differently and then have a different kind of gene in the middle. Traditional methods struggle to get this area right.4
Correcting Assemblies with BAC Sequences
To deal with bacterial artificial chromosome (BAC) sequencing issues, soybean researchers used a special approach. They looked at BAC clones and sequenced certain ones that matched with a CHS cDNA. This method helped them see how multiple CHS genes fit together in a 10.9 kilobase section. It shed light on the complex structure of the area.4
| Key Statistics | Data |
|---|---|
| Repeat content in maize genome | 80%4 |
| Genome duplication in Arabidopsis and soybean progenitors | Regions of genome duplication have occurred4 |
| Repeated DNA regions and genome assembly issues | Repeated DNA regions often lead to mis‐assemblies and gaps in sequenced genomes, affecting the assembly of short next‐generation sequence (NGS) reads4 |
| CHS genes identified through conventional cloning and sequencing | At least nine CHS family members have been identified4 |
| Structure of the 27-kb region in the soybean genome | An unusual 27‐kb region in the soybean genome consists of two identical 10.91‐kb inverted repeats of CHS1, CHS3, CHS4 and CHS4, CHS3, CHS1 separated by 5.71 kb of intervening sequence that codes for one hypothetical protein4 |
Mechanisms of Non Allelic Homologous Recombination
Data indicates how non allelic homologous recombination (NAHR) works. It swaps genetic material between areas with similar sequences. These areas might not be on the same chromosome or site. This exchange can happen during meiosis when similar chromosomes pair up.2 This pairing error can cause a crossover of genetic materials, leading to changes in DNA structure. If this process involves duplicates created over time, it may lead to deletions or additions of genetic material. This could cause rare genetic conditions by affecting genes negatively or positively in the region.
NAHR might happen between duplicated DNA sequences up to 300 kbp in size.5 To find these changes, scientists use optical genome mapping. This method is good at spotting big changes in DNA, like missing or extra parts.5 Such changes are a big part of how our DNA is different from each other. By using optical mapping, experts have found genetic issues in diseases like cancer and muscular dystrophy. This technique, along with mapping the entire genome, makes it easier to identify these big changes.
A detailed study focused on 44 people to look at NAHR’s effects. It found that 109 of the 324 possible genetic swap spots showed deletions or additions.3 This work also inferred over 1,000 genetic swapping events among people’s DNA. These events were often new and not seen before in similar studies. This shows how unique each person’s genome can be in these swapping cases.3 Also, most of these genetic swap locations had not been well noticed by previous methods. The study used a smart computer model to find these hidden places of genetic change. This approach made it possible to see new genetic swaps that other methods missed.
Genomic Disorders and NAHR
Non allelic homologous recombination (NAHR) plays a key role in various genomic disorders in people. It can cause changes in chromosomes, like deletions or duplications. These changes mess with how genes work, leading to health issues.3
Role of NAHR in Human Disease Traits
About 10% to 22% of genetic changes in people are because of NAHR. In a study with 44 people, over 1,000 NAHR events were found. These findings show NAHR is significant in human genetics.3 It was also noted that the chance of NAHR happening can relate to a person’s background. This suggests genes might influence NAHR.3
Examples of NAHR-Mediated Disorders
Smith-Magenis syndrome, Potocki-Lupski syndrome, and 17p11.2 deletions are linked to NAHR.1 The process involves recombination of similar gene sections. For example, a condition where some genes are missing is caused by this. Additionally, in some cases, genes can end up duplicated due to similar DNA sequences.1
Such deletions and duplications in certain areas can be common when specific DNA pieces are involved. This shows how complex and varied the effects of NAHR can be.1
| Genomic Disorder | NAHR-Mediated Mechanism | References |
|---|---|---|
| Smith-Magenis syndrome | Homologous recombination of flanking repeat gene clusters | 1 |
| Potocki-Lupski syndrome | Uncommon recurrent duplications | 1 |
| 17p11.2 deletions | Homologous and non-homologous mechanisms | 1 |
Recombination Hotspots and NAHR
Non allelic homologous recombination (NAHR) is affected by recombination hotspots. These are areas with more meiotic recombination than others nearby.1
PRDM9 and Meiotic Recombination Hotspots
The PRDM9 gene is vital for creating recombination hotspots in meiosis. Research indicates PRDM9 directs where and how these hotspots work, affecting how meiotic recombination happens across the genome.1 So, changes in PRDM9 can alter recombination landscapes, changing how NAHR and chromosomal alterations occur.
A common sequence motif has been found in recombination hotspots.1 This discovery helps us understand how NAHR works at a molecular level. Knowing this sheds light on the connection between NAHR, genome structure, and genetic diseases.
NAHR in Human Genome and Fertility
Non allelic homologous recombination (NAHR) plays a part in male infertility. It causes genetic changes when chromosomes swap material incorrectly. This leads to issues in how chromosomes pair up and separate during cell division, possibly making eggs or sperm abnormal. This can then cause fertility problems.6
Role of NAHR in Male Infertility
NAHR is a key factor in male infertility, shows research. For instance, a study in India found that nearly a third of infertile men had deletion mutations. These deletions were mainly due to NAHR. They specifically looked at deletions in regions known as AZF, critical for sperm production.7 The study found various kinds of deletions in these regions, showing the importance of NAHR in male infertility.7
Mechanisms of NAHR in Meiotic Recombination
Scientists have researched a lot about how NAHR operates at a genetic level. They’ve uncovered how chromosomes swap parts in a process called recombination. They compare human and mouse studies with yeast to understand genetic crossing-over better. This information helps in studying how genes evolve.6 There’s even a specific DNA sequence identified in many recombination sites. This sequence appears in over 40% of the recombination hotspots. It gives clues about the specific steps in these processes.6
Future Perspectives and Challenges
Scientists are learning more about non allelic homologous recombination (NAHR) as time goes on. The work of Conrad DF et al.8 and Korbel JO et al.8, along with the 1000 Genomes Project Consortium8, is key here. They studied how our genes can change over time.8
Yet, we still need better math models to understand how genes that are similar can switch places. This issue was pointed out in recent studies9.
CRISPR/Cas910 and similar cutting-edge tools are speeding up our ability to change genes. They are also helping us see how NAHR affects different living things.10 Studying NAHR could shed light on gene-related diseases and improve our health. Stankiewicz P, Lupski JR8, and Carvalho CM et al.8 have looked into this.
Recently, a major milestone was reached with the full mapping of a human genome’s ends9. The Human Pangenome Reference Consortium is also hard at work creating detailed gene maps9. These efforts promise to show us more about how our genes can change.
Scientists will keep digging into how NAHR works and its effects on us. They’re keen to use the latest gene tech to better understand and maybe control NAHR. Meeting these future perspectives and challenges is key to moving our research forward. It could lead to exciting new ways to treat diseases and use gene tech.
Source Links
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3188830/
- https://en.wikipedia.org/wiki/Non-allelic_homologous_recombination
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0633-1
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6710647/
- https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/non-allelic-homologous-recombination
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3073813/
- https://www.nature.com/articles/s41598-019-42690-0
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4425883/
- https://almob.biomedcentral.com/articles/10.1186/s13015-023-00241-3
- https://www.nature.com/articles/s41392-019-0089-y